Summary
- As of 13 January 2022, the UP Philippine Genome Center, in cooperation with the Department of Health Epidemiology Bureau and the UP National Institutes of Health, has detected 517 cases of the SARS-CoV-2 Omicron variant in the country. Of these, 182 were from international travelers while 335 were local transmissions.
- Omicron cases were initially detected early in December 2021 and continue to sharply increase, with almost all sequenced samples collected during the first days of January 2022 classified under this variant. This rapid increase in Omicron cases may be attributed to the high transmissibility and immune evasion of this variant, similar to what has been observed in other countries.
- Analysis of genome sequences reveal that the majority of Omicron cases in international travelers did not appear to have onward transmission of the virus. However, a number of undetected cases and quarantine breaches could have led to the entry and rapid local spread of the said variant of concern.
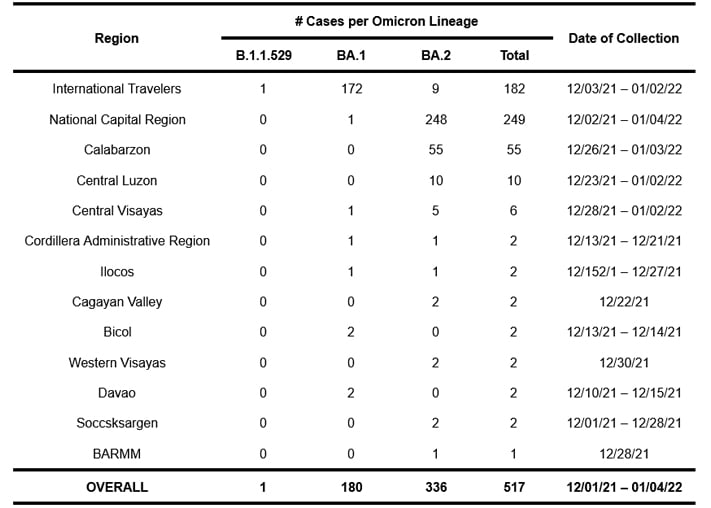
- Local cases of the Omicron variant mostly belong to Omicron sublineage BA.2 and have already been detected in 12 regions of the country, with the majority of cases detected in NCR and Calabarzon.
- Phylodynamic patterns from the local Omicron sequences suggest exponential and rapid spread of the virus – consistent with the occurrence of community transmission. Clustering of cases from various regions of the country was also observed, hinting at porous interzonal borders.
- The detection of two distinct mutations in local samples suggests that a majority of the local Omicron cases are being sustained and driven through community transmission of the circulating lineages.
- The Omicron variant was detected in a wide range of age groups similar to previous variants. Omicron was also detected in fully vaccinated individuals though the disease presentation was mostly asymptomatic to mild. Based on available case information, a strong correlation between the Omicron sublineage and the presentation of symptoms was also observed – with initial data suggesting symptoms to more likely be observed in BA.2 cases than in BA.1 cases. Further analysis need to be done to determine whether this observation is a consequence of sampling bias or the innate characteristic of the variant.
- Compliance to health protocols is strongly encouraged even for fully vaccinated or previously infected individuals. Wider sampling of cases for genomic biosurveillance is necessary to have a more accurate picture of the distribution and spread of this variant in the country.
Technical Report
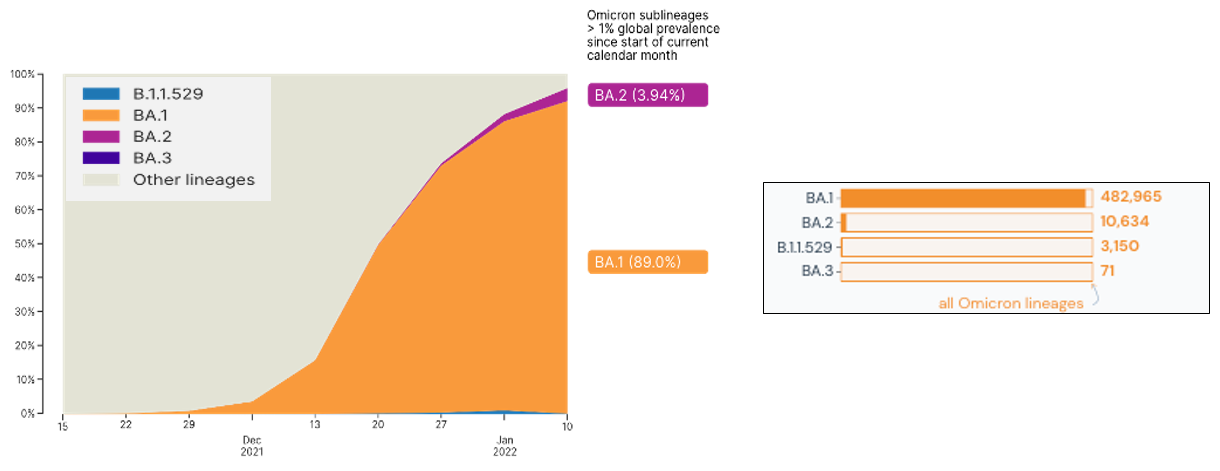
The Omicron variant was first identified in Botswana and South Africa, emerging at the tail end of the Delta surge. At present, cases of the Omicron variant continue to increase and have been reported in over 118 countries with over 496,000 sequences in the combined Omicron lineages shared in global databases. This variant, with its more than 30 mutations in the Spike region is hypothesized to be over twice as infectious and twice more likely to escape current vaccines than the Delta variant. Currently, Omicron consists of four (4) phylogenetic lineages: the main lineage B.1.1.529 and its three (3) sublineages: BA.1, BA.2, and BA.3. Based on global statistics, BA.1 is the predominant Omicron lineage being detected worldwide, but a recent increase in the proportion of BA.2 cases has also been observed (Figure 1).
Figure 1. Global proportion on lineages associated with the Omicron variant. Image on the left is from GISAID.org (gisaid.org/hcov-19-analysis-update) while data on the right is from outbreak.info, accessed on January 20, 2022.
Spatial and temporal distribution of Omicron cases
To date, 517 samples sequenced and analyzed at the UP Philippine Genome Center have been classified as belonging to the Omicron variant based on their genome sequences. Of these cases, 182 were detected from International Travelers (ITs), whereas 335 were local transmissions spanning 12 regions in the country, with the majority coming from the National Capital Region (NCR). Table 1 lists the regions where the Omicron variant has been detected. As only a small percentage of all COVID-19 cases are submitted to the UP PGC for sequencing and analysis, note possible bias in sampling with uneven rates across locations and selective sampling.
The temporal distribution of these cases (Figure 2) show that a steep increase in the proportion of detected Omicron samples occurred towards the latter half of December 2021. This trend continued until the start of 2022, with almost all samples analyzed so far this year classified under this variant. The rapid rise in the number of local Omicron cases may be attributed to the high transmissibility and immune evasion of this variant, similar to what has been observed in other countries.
Figure 2. Raw counts (top) and proportions (bottom) of SARS-CoV-2 variants detected from local samples collected since October 2021, shown on a weekly interval. The Delta variant surge in the country has already reached its peak during this period and the actual number of confirmed COVID-19 cases dwindled until the third week of December 2021 (reflected in the ribbon chart). The earliest cases of Omicron variant were detected early in December and a steep increase in the proportion of the said variant can be observed since, with Omicron rapidly overtaking Delta as the dominant local SARS-CoV-2 variant at the beginning of 2022. (Note: possible sampling bias from uneven rates of sequencing across locations and may not represent the true prevalence of the variants in the population).
Phylodynamic analysis
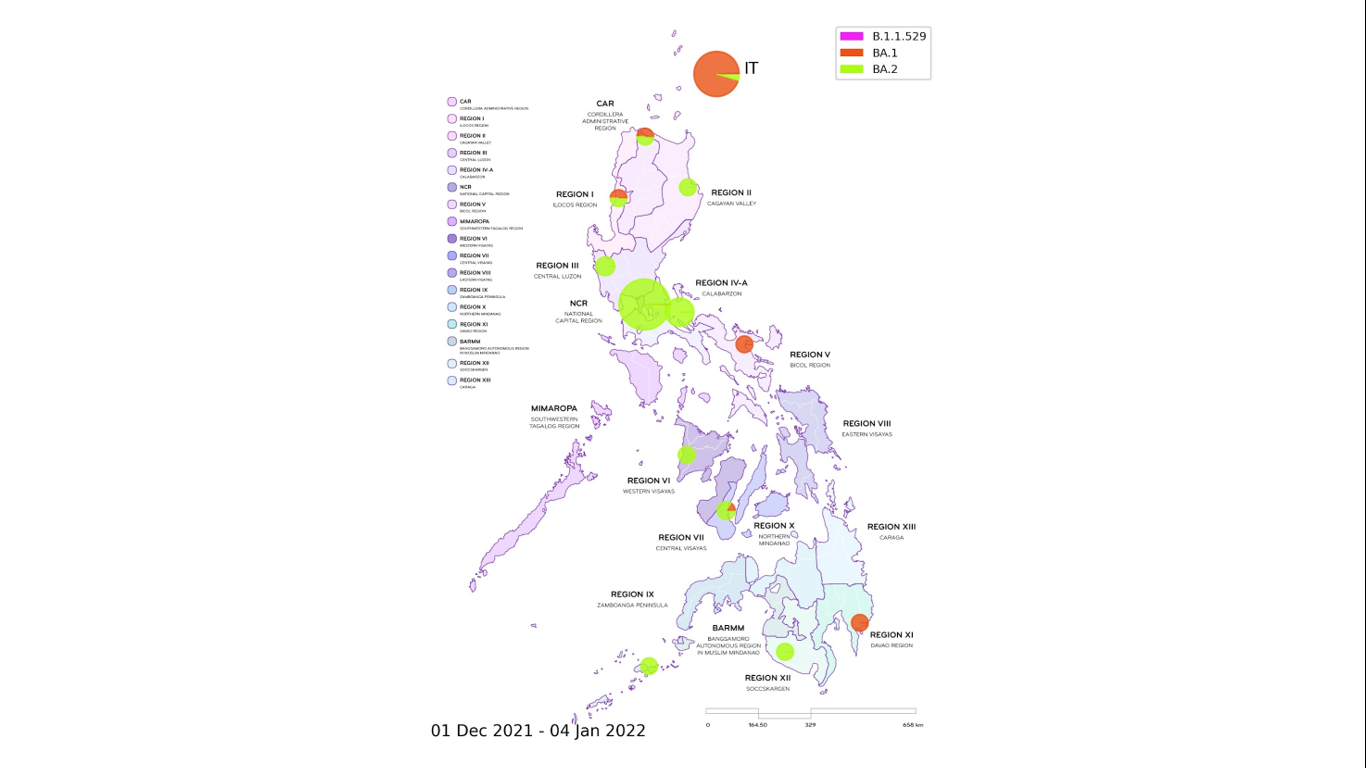
Figure 3. Distribution map of locally-detected Omicron cases. The variant has been detected in 12 different regions in the country, with most of the observed local Omicron cases classified as belonging to the BA.2 lineage, whereas those from international travelers (IT) were mainly under the BA.1 lineage. For local cases, Region is based on the recorded patient address whenever available. If no information is provided, the collecting institution region is used. (Note: possible sampling bias from uneven rates of sequencing across locations and may not represent the true prevalence of the variants in the population).
The distribution map of all detected Omicron variant cases is shown in Figure 3. Local cases of Omicron have been observed in most regions in the country except for MIMAROPA, Eastern Visayas, Zamboanga, and Northern Mindanao where only a small number of samples have been submitted for whole genome sequencing.
The strong correlation between the observed lineage of Omicron cases and the case type (whether local or IT) – with the majority of cases from international travelers classified as belonging to the BA.1 lineage while local transmissions mostly fall under the BA.2 lineage – suggests that many of the detected Omicron cases in international travelers did not have onward transmission of the virus. However, a number of undetected cases and quarantine breaches could have led to the entry and rapid local spread of the said variant of concern.
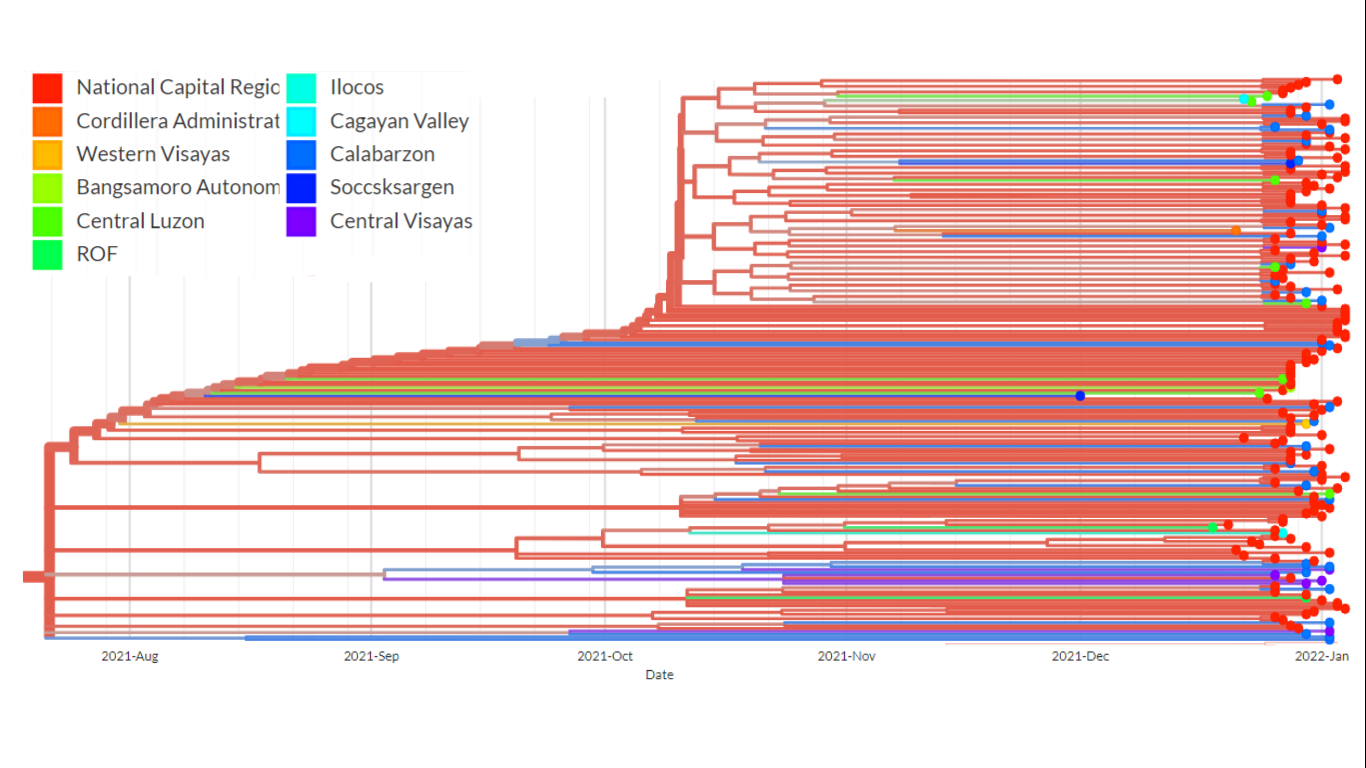
Figure 4. A pruned branch from the time-scaled phylogenetic tree of the Omicron samples. This particular cluster shows the presence of relatively long branches leading to terminal nodes, a phylodynamic pattern suggestive of exponential and rapid transmission. Weak spatial structure can also be observed as samples from different regions forming close clusters.
A close look at the topology of certain clusters in the time-scaled phylogenetic tree (Figure 4) shows the presence of long branches leading to terminal nodes – a phylodynamic pattern that is consistent with exponential growth and spread of the virus. This observation provide evidence that community transmission, particularly of the BA.2 lineage of Omicron, is already occurring. In addition, many of the phylogenetic clusters were observed to have weak spatial structure, suggesting the presence of inter-regional transmissions and porous borders.
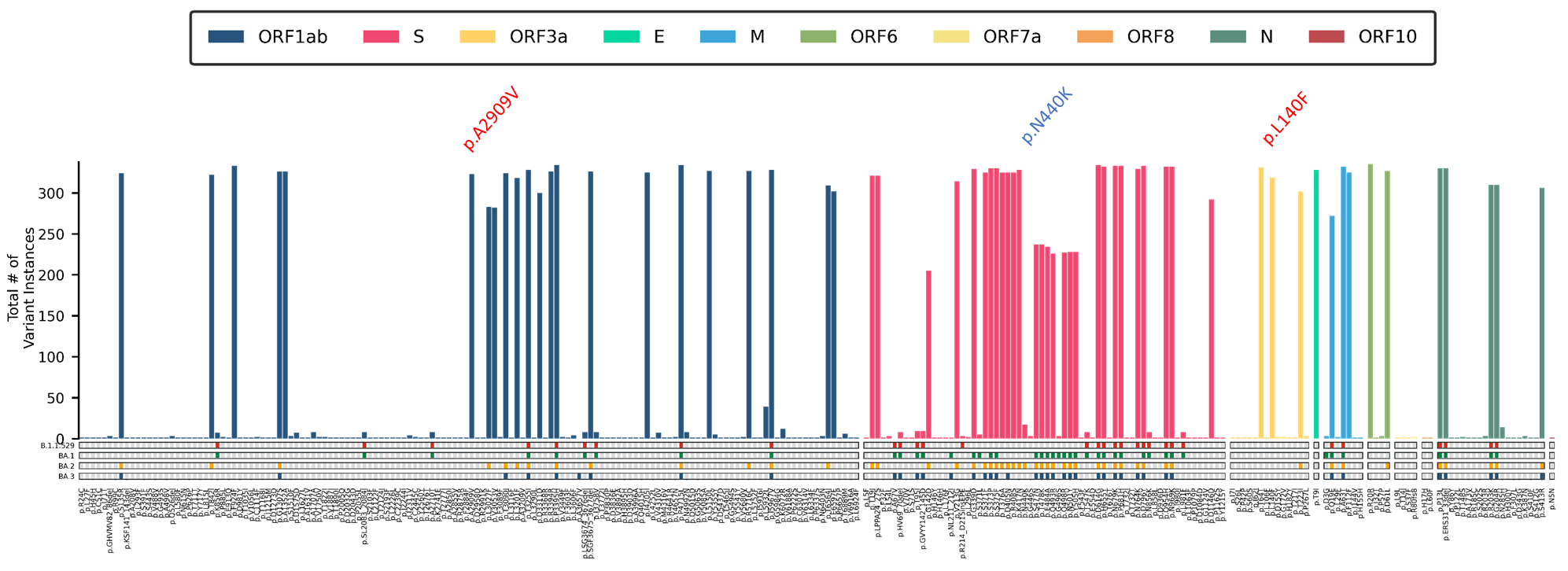
Among the local Omicron samples, a locally distinct mutation profile is observed to be developing (Figure 5). These samples appear to be carrying two (2) mutations that do not appear in the global mutation profile for any of the Omicron sublineages: A2909V in the ORF1ab gene, and L140F in the ORF3a gene. Additionally, one global signature Omicron mutation (S: N440K) is noticeably absent. The homogeneity of these distinct features in the local population, alongside the phylogenetic analysis, further suggests that a majority of the local Omicron cases are being sustained and driven through community transmission of the circulating lineages.
Figure 5. Mutation Profile of 335 local Omicron cases detected in the country. These include two (2) mutations that are not key mutations in any other Omicron lineage (red), and 1 key BA.2 spike protein mutation that is distinctly absent (blue).
Age and sex distribution and clinical presentation
The age distribution of individuals detected to carry the Omicron variant appears to be wide, with patients ranging from infants (<1 year old) to over 85 years old. The majority of these cases were detected in the working segment of the population (20 to 40 years old), similarly affecting male and female individuals. For cases with disease severity information, no significant difference in severity was observed among males and females (data not shown).
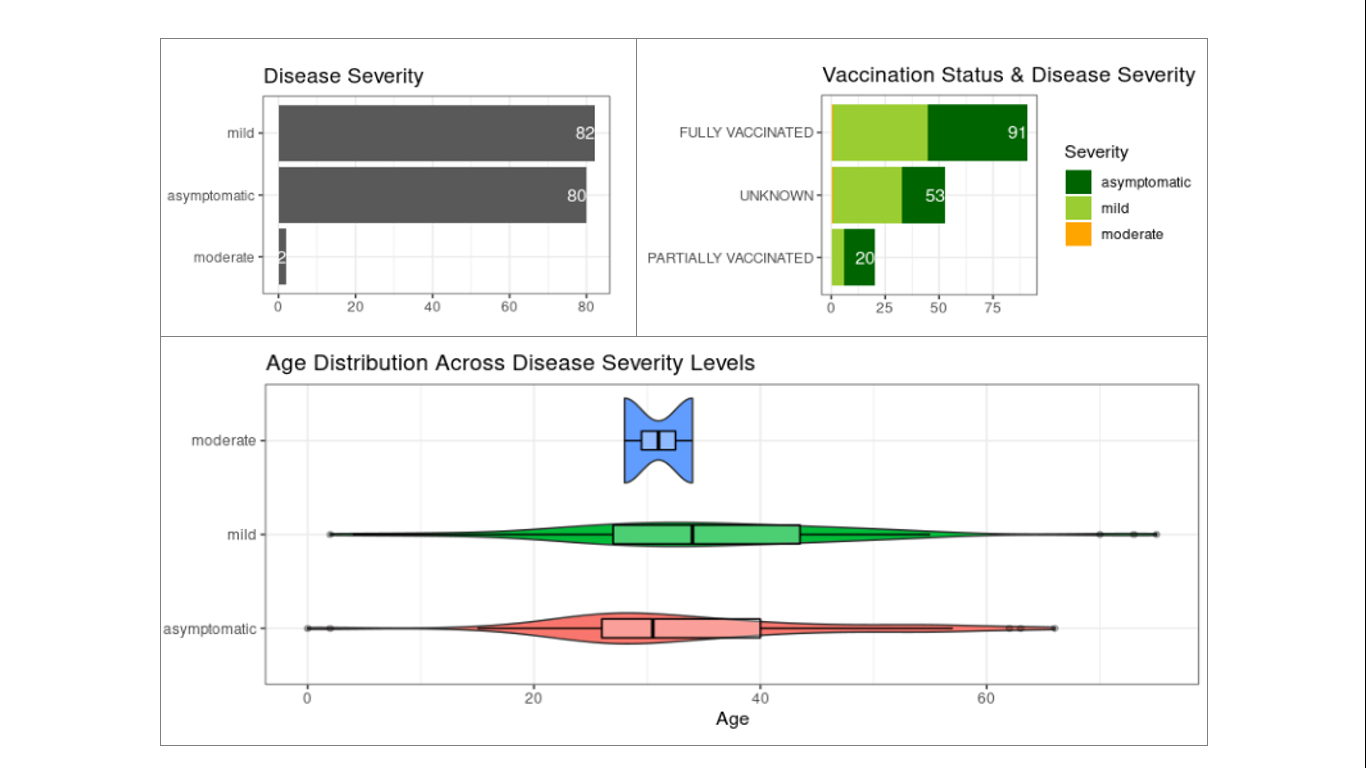
Figure 6. Disease severity status of detected Omicron cases. Disease severity data are based on 164 cases with available information from the total 517 samples. Most of these cases were tagged as asymptomatic or mild (top left panel) including from fully vaccinated individuals (top right panel). No significant correlation was observed between disease severity and age, although the distribution of cases with mild symptoms appear to be slightly shifted towards the older age group.
Most of the detected Omicron cases with available disease severity information (164 out of 517 samples) were asymptomatic to mild, with only two (2) moderate cases (Figure 6). Interestingly, a substantial number of infections occurred in fully vaccinated individuals with many of the cases reported to not develop severe symptoms. This observation reflects the immune evasion capability of the Omicron variant and the protection offered by full vaccination in preventing development of severe to critical symptoms among the fully vaccinated.
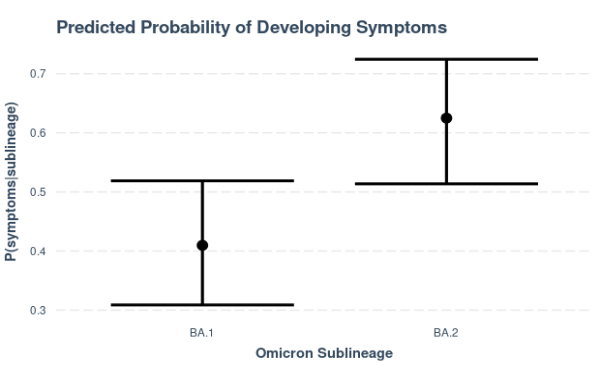
Figure 7. Predicted probabilities of having symptoms, estimated with a binomial regression model with sublineage as the sole predictor. Cases belonging to the BA.2 sublineage (n=80) are predicted to be more likely to develop symptoms (P(symptoms|BA.2) = 0.62) than those classified under the BA.1 sublineage (n=83). Caution is noted in the interpretation of these results due to limited severity information and potential sampling bias of local cases included in genome sequencing, most of which belong to the BA.2 sublineage.
The available data so far also suggest that there is a strong association (p𝜒2=0.006) between the detected Omicron sublineage and the presentation of disease symptoms. In particular, the probability of developing symptoms is estimated to be significantly higher in cases belonging to the BA.2 sublineage compared to those classified under the BA.1 sublineage. This observation is based on the relatively low proportion of Omicron samples with recorded disease severity information and it remains to be seen whether this will hold true with a bigger sample population. Caution, too, is noted in the interpretation of these initial results due to potential sampling bias of local cases included for genome sequencing.
Recommendations
In light of these recent findings, vaccination and compliance to health protocols is strongly recommended for everyone. Wider sampling of cases for genomic biosurveillance is likewise necessary to have a more accurate picture of the distribution and spread of this variant in the country.
(This article was first published in the PGC Website on January 20, 2022)